Science
Researchers Master Control of Proteins in Living Animals
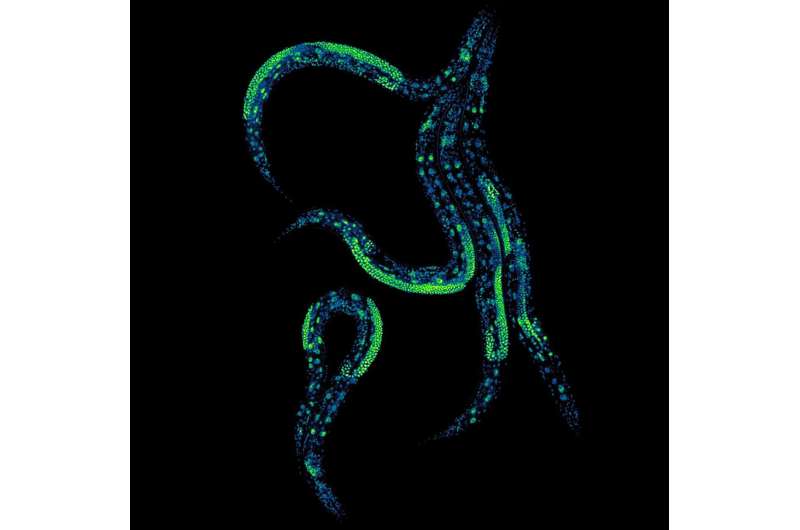
Researchers have pioneered a groundbreaking method that enables precise control of protein levels in living animals throughout their lifespan. This innovative technique not only enhances the understanding of biological processes but also opens new avenues for investigating aging and disease. The findings, led by scientists from the Center for Genomic Regulation in Barcelona and the University of Cambridge, were published on December 12, 2025, in the journal Nature Communications.
Revolutionizing Protein Control
The research team successfully manipulated protein levels in the intestines and neurons of the nematode worm Caenorhabditis elegans. Their ability to adjust protein concentrations with precision marks a significant advancement in molecular biology. As noted by Dr. Nicholas Stroustrup, a researcher at the Center for Genomic Regulation, “No protein acts alone. Our new approach lets us study how multiple proteins in different tissues cooperate to control how the body functions and ages.”
This method addresses a critical limitation of prior techniques, which often focused on binary on/off switches for protein activity. Traditional approaches could not adequately separate the effects of proteins across different body parts or provide the nuanced control necessary for understanding complex biological interactions.
Dual-Channel AID System Explained
The innovation stems from an adaptation of the auxin-inducible degron system (AID), originally developed in plant biology. In this system, proteins are tagged with a degron, a marker that prompts their degradation in the presence of auxin, a plant hormone. The researchers engineered a “dual-channel” version of this system, allowing them to control protein levels more flexibly.
By employing two distinct TIR1 enzymes, each activated by different auxin compounds, the researchers can manage protein levels in various tissues independently. This means they can adjust the same protein in both the intestine and neurons or regulate different proteins simultaneously. The study’s authors emphasized that achieving this level of control required overcoming significant engineering challenges, including ensuring that the two synthetic switches did not interfere with one another.
The versatility of this method extends to reproductive tissues, which have historically posed difficulties for AID systems. The research team adapted their approach to ensure functionality in these areas, providing a comprehensive tool for studying systemic biological processes.
Dr. Jeremy Vicencio, a postdoctoral researcher and co-author of the study, remarked on the potential impact of this innovation. “It’s a powerful tool that we hope will open up new possibilities for biologists everywhere,” he stated.
The implications of this research are profound, as it will enable scientists to undertake experiments previously deemed impossible. By understanding how varying protein levels contribute to health and aging, researchers can begin to unravel the intricate molecular networks that govern these processes. This advancement may ultimately lead to novel strategies for addressing age-related diseases and improving overall health.
The study represents a significant leap forward in the field of molecular biology, offering tools that could reshape research methodologies and deepen our understanding of fundamental biological principles. As scientists continue to explore the nuances of protein interactions, the insights gained may pave the way for innovative therapeutic approaches in the future.
-
 Science4 weeks ago
Science4 weeks agoALMA Discovers Companion Orbiting Giant Red Star π 1 Gruis
-

 Politics2 months ago
Politics2 months agoSEVENTEEN’s Mingyu Faces Backlash Over Alcohol Incident at Concert
-

 Top Stories2 months ago
Top Stories2 months agoNew ‘Star Trek: Voyager’ Game Demo Released, Players Test Limits
-

 World2 months ago
World2 months agoGlobal Air Forces Ranked by Annual Defense Budgets in 2025
-

 World2 months ago
World2 months agoMass Production of F-35 Fighter Jet Drives Down Costs
-
 World2 months ago
World2 months agoElectrification Challenges Demand Advanced Multiphysics Modeling
-

 Business2 months ago
Business2 months agoGold Investment Surge: Top Mutual Funds and ETF Alternatives
-

 Science2 months ago
Science2 months agoTime Crystals Revolutionize Quantum Computing Potential
-

 Top Stories2 months ago
Top Stories2 months agoDirecTV to Launch AI-Driven Ads with User Likenesses in 2026
-

 Entertainment2 months ago
Entertainment2 months agoFreeport Art Gallery Transforms Waste into Creative Masterpieces
-

 Business2 months ago
Business2 months agoUS Government Denies Coal Lease Bid, Impacting Industry Revival Efforts
-

 Health2 months ago
Health2 months agoGavin Newsom Critiques Trump’s Health and National Guard Plans